Reference-Based Multiple Imputation
A comparison of Reference-Based Multiple Imputation between R and SAS
A recent PHUSE CAMIS contribution (in collaboration with PSI AIMS SIG) explored the implementation of reference-based multiple imputation in R and SAS, and a comparison of both. The focus was on continuous longitudinal endpoints.
Multiple imputation is a common method that statisticians use to handle missing data in biostatistics. Instead of discarding incomplete cases or using single imputation methods that underestimate variability, multiple imputation creates several plausible datasets by replacing missing values using an appropriate stochastic model based on observed data. Each complete dataset is analysed using the specified model, and results are combined using Rubin’s rules to account for both within- and between-imputation variability. This ensures valid inference under the so-called missing-at-random (MAR) assumption.
Carpenter et al (2013) introduced reference-based multiple imputation, particularly for sensitivity analyses of missing data results under missing not-at-random (MNAR) assumptions. Unlike standard multiple imputation, which predicts missing values based on the participant’s own treatment group, reference-based multiple imputation uses data from a reference group (e.g., the control arm) to impute post-dropout values for participants in the experimental arm. The idea is to reflect clinically plausible scenarios, such as assuming that after discontinuation of treatment, a patient’s outcomes would behave as if they were on the reference treatment (e.g., the control arm).
In R, the rbmi package by Gower-Page et al (2022) was used. In rbmi, the reference-based multiple imputation approach is implemented by fitting a Bayesian full multivariate normal repeated measures model using MCMC and then drawing posterior samples to perform imputations. The following reference-based multiple imputation approaches are available in rbmi: copy increments from reference (CIR), copy reference (CR) and jump to reference (JR).
In SAS, five macros (implemented by the London School of Hygiene & Tropical Medicine) are available to perform reference-based multiple imputation. The code consists of a set of 5 macros, and the approach is implemented as described above for the rbmi package, also with the options of CIR, CR, and JR (specified as J2R within the SAS five macros).
The PHUSE CAMIS project provides a detailed step-by-step overview on the implementation of reference-based multiple imputation in R rbmi and SAS five macros using an example dataset (full details here: R vs SAS Reference-Based Multiple Imputation (joint modelling): Continuous Data). A comparison of analysis results from both software revealed that results were matching completely (up to some small Monte Carlo error given the randomness in multiple imputations). Standard multiple imputation and three reference-based multiple imputation approaches (CIR, CR, JR) were compared for datasets with different numbers of imputations (i.e., 500, 2000 and 5000).The figure below compares, for an example dataset, the contrast estimates (and associated 95% confidence interval) between R and SAS.
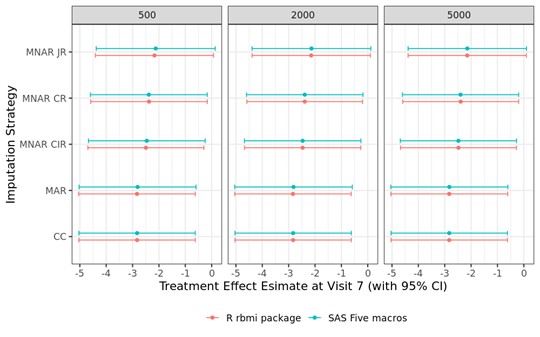
CC = Complete Case; MAR = Missing at Random; MNAR = Missing not at Random, CIR = Copy Increments in Reference; CR = Copy Reference; JR = Jump to Reference.
References:
Carpenter JR, Roger JH & Kenward MG (2013). Analysis of Longitudinal Trials with Protocol Deviation: A Framework for Relevant, Accessible Assumptions, and Inference via MI. Journal of Biopharmaceutical Statistics 23: 1352-1371.
Gower-Page C, Noci A & Wolbers M (2022). rbmi: A R package for standard and reference-based multiple imputation methods. Journal of Open Source Software 7(74): 4251. https://www.lshtm.ac.uk/research/centres-projects-groups/missing-data#dia-missing-data